publications
publications by categories in reversed chronological order.
2025
- NLDL
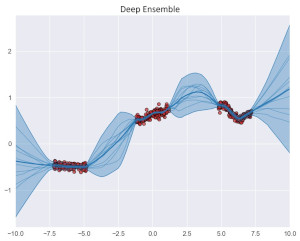 Hallucination Detection in LLMs: Fast and Memory-Efficient Finetuned ModelsGabriel Y. Arteaga, Thomas B. Schön, and Nicolas PielawskiIn Proceedings of the 6th Northern Lights Deep Learning Conference (NLDL), 07–09 jan 2025
Hallucination Detection in LLMs: Fast and Memory-Efficient Finetuned ModelsGabriel Y. Arteaga, Thomas B. Schön, and Nicolas PielawskiIn Proceedings of the 6th Northern Lights Deep Learning Conference (NLDL), 07–09 jan 2025Uncertainty estimation is a necessary component when implementing AI in high-risk settings, such as autonomous cars, medicine, or insurances. Large Language Models (LLMs) have seen a surge in popularity in recent years, but they are subject to hallucinations, which may cause serious harm in high-risk settings. Despite their success, LLMs are expensive to train and run: they need a large amount of computations and memory, preventing the use of ensembling methods in practice. In this work, we present a novel method that allows for fast and memory-friendly training of LLM ensembles. We show that the resulting ensembles can detect hallucinations and are a viable approach in practice as only one GPU is needed for training and inference.
@inproceedings{hallucinationDetection2025, title = {Hallucination Detection in {LLM}s: Fast and Memory-Efficient Finetuned Models}, author = {Arteaga, Gabriel Y. and Sch{\"o}n, Thomas B. and Pielawski, Nicolas}, booktitle = {Proceedings of the 6th Northern Lights Deep Learning Conference (NLDL)}, pages = {1--15}, year = {2025}, editor = {Lutchyn, Tetiana and Ramírez Rivera, Adín and Ricaud, Benjamin}, volume = {265}, series = {Proceedings of Machine Learning Research}, month = {07--09 Jan}, publisher = {PMLR}, url = {https://proceedings.mlr.press/v265/arteaga25a.html}, }
2023
- Heliyon
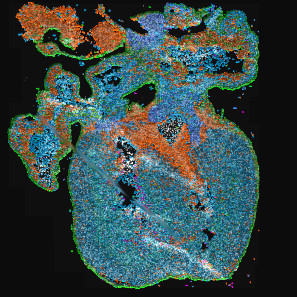 TissUUmaps 3: Improvements in interactive visualization, exploration, and quality assessment of large-scale spatial omics dataNicolas Pielawski, Axel Andersson, Christophe Avenel, and 6 more authorsHeliyon, 07–09 jan 2023
TissUUmaps 3: Improvements in interactive visualization, exploration, and quality assessment of large-scale spatial omics dataNicolas Pielawski, Axel Andersson, Christophe Avenel, and 6 more authorsHeliyon, 07–09 jan 2023Background and objectives Spatially resolved techniques for exploring the molecular landscape of tissue samples, such as spatial transcriptomics, often result in millions of data points and images too large to view on a regular desktop computer, limiting the possibilities in visual interactive data exploration. TissUUmaps is a free, open-source browser-based tool for GPU-accelerated visualization and interactive exploration of 107+ data points overlaying tissue samples. Methods Herein we describe how TissUUmaps 3 provides instant multiresolution image viewing and can be customized, shared, and also integrated into Jupyter Notebooks. We introduce new modules where users can visualize markers and regions, explore spatial statistics, perform quantitative analyses of tissue morphology, and assess the quality of decoding in situ transcriptomics data. Results We show that thanks to targeted optimizations the time and cost associated with interactive data exploration were reduced, enabling TissUUmaps 3 to handle the scale of today’s spatial transcriptomics methods. Conclusion TissUUmaps 3 provides significantly improved performance for large multiplex datasets as compared to previous versions. We envision TissUUmaps to contribute to broader dissemination and flexible sharing of largescale spatial omics data.
@article{TissUUmaps2023, title = {TissUUmaps 3: Improvements in interactive visualization, exploration, and quality assessment of large-scale spatial omics data}, journal = {Heliyon}, volume = {9}, number = {5}, pages = {e15306}, year = {2023}, issn = {2405-8440}, doi = {10.1016/j.heliyon.2023.e15306}, url = {https://www.sciencedirect.com/science/article/pii/S2405844023025136}, author = {Pielawski, Nicolas and Andersson, Axel and Avenel, Christophe and Behanova, Andrea and Chelebian, Eduard and Klemm, Anna and Nysjö, Fredrik and Solorzano, Leslie and Wählby, Carolina}, keywords = {Interactive visualization, Spatial omics, Spatial transcriptomics}, } -
 Learning-based prediction, representation, and multimodal registration for bioimage processingNicolas PielawskiUppsala University, Mar 2023
Learning-based prediction, representation, and multimodal registration for bioimage processingNicolas PielawskiUppsala University, Mar 2023Microscopy and imaging are essential to understanding and exploring biology. Modern staining and imaging techniques generate large amounts of data resulting in the need for automated analysis approaches. Many earlier approaches relied on handcrafted feature extractors, while today’s deep-learning-based methods open up new ways to analyze data automatically. Deep learning has become popular in bioimage processing as it can extract high-level features describing image content (Paper III). The work in this thesis explores various aspects and limitations of machine learning and deep learning with applications in biology. Learning-based methods have generalization issues on out-of-distribution data points, and methods such as uncertainty estimation (Paper II) and visual quality control (Paper V) can provide ways to mitigate those issues. Furthermore, deep learning methods often require large amounts of data during training. Here the focus is on optimizing deep learning methods to meet current computational capabilities and handle the increasing volume and size of data (Paper I). Model uncertainty and data augmentation techniques are also explored (Papers II and III). This thesis is split into chapters describing the main components of cell biology, microscopy imaging, and the mathematical and machine-learning theories to give readers an introduction to biomedical image processing. The main contributions of this thesis are deep-learning methods for reconstructing patch-based segmentation (Paper I) and pixel regression of traction force images (Paper II), followed by methods for aligning images from different sensors in a common coordinate system (named multimodal image registration) using representation learning (Paper III) and Bayesian optimization (Paper IV). Finally, the thesis introduces TissUUmaps 3, a tool for visualizing multiplexed spatial transcriptomics data (Paper V). These contributions provide methods and tools detailing how to apply mathematical frameworks and machine-learning theory to biology, giving us concrete tools to improve our understanding of complex biological processes.
@phdthesis{pielawski2023, author = {Pielawski, Nicolas}, title = {Learning-based prediction, representation, and multimodal registration for bioimage processing}, school = {Uppsala University}, year = {2023}, month = mar, address = {Ångström Laboratory, Uppsala, Sweden}, url = {https://urn.kb.se/resolve?urn=urn:nbn:se:uu:diva-497143}, isbn = {978-91-513-1725-0}, }
2020
- PLOS ONE
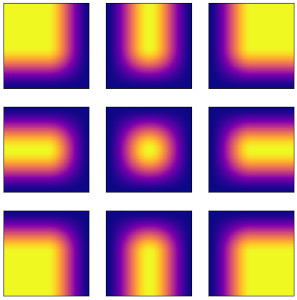 Introducing Hann windows for reducing edge-effects in patch-based image segmentationNicolas Pielawski and Carolina WählbyPLOS ONE, Mar 2020
Introducing Hann windows for reducing edge-effects in patch-based image segmentationNicolas Pielawski and Carolina WählbyPLOS ONE, Mar 2020There is a limitation in the size of an image that can be processed using computationally demanding methods such as e.g. Convolutional Neural Networks (CNNs). Furthermore, many networks are designed to work with a pre-determined fixed image size. Some imaging modalities—notably biological and medical—can result in images up to a few gigapixels in size, meaning that they have to be divided into smaller parts, or patches, for processing. However, when performing pixel classification, this may lead to undesirable artefacts, such as edge effects in the final re-combined image. We introduce windowing methods from signal processing to effectively reduce such edge effects. With the assumption that the central part of an image patch often holds richer contextual information than its sides and corners, we reconstruct the prediction by overlapping patches that are being weighted depending on 2-dimensional windows. We compare the results of simple averaging and four different windows: Hann, Bartlett-Hann, Triangular and a recently proposed window by Cui et al., and show that the cosine-based Hann window achieves the best improvement as measured by the Structural Similarity Index (SSIM). We also apply the Dice score to show that classification errors close to patch edges are reduced. The proposed windowing method can be used together with any CNN model for segmentation without any modification and significantly improves network predictions.
@article{hannwindows2020, doi = {10.1371/journal.pone.0229839}, author = {Pielawski, Nicolas and Wählby, Carolina}, journal = {PLOS ONE}, publisher = {Public Library of Science}, title = {Introducing Hann windows for reducing edge-effects in patch-based image segmentation}, year = {2020}, month = mar, volume = {15}, url = {https://doi.org/10.1371/journal.pone.0229839}, pages = {1-11}, number = {3}, } - ISBI
 In Silico Prediction of Cell Traction ForcesNicolas Pielawski, Jianjiang Hu, Staffan Strömblad, and 1 more authorIn 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Mar 2020
In Silico Prediction of Cell Traction ForcesNicolas Pielawski, Jianjiang Hu, Staffan Strömblad, and 1 more authorIn 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Mar 2020Traction Force Microscopy (TFM) is a technique used to determine the tensions that a biological cell conveys to the underlying surface. Typically, TFM requires culturing cells on gels with fluorescent beads, followed by bead displacement calculations. We present a new method allowing to predict those forces from a regular fluorescent image of the cell. Using Deep Learning, we trained a Bayesian Neural Network adapted for pixel regression of the forces and show that it generalises on different cells of the same strain. The predicted forces are computed along with an approximated uncertainty, which shows whether the prediction is trustworthy or not. Using the proposed method could help estimating forces when calculating non-trivial bead displacements and can also free one of the fluorescent channels of the microscope. Code is available at https://github.com/wahlby-lab/InSilicoTFM.
@inproceedings{insilicotfm2020, author = {Pielawski, Nicolas and Hu, Jianjiang and Strömblad, Staffan and Wählby, Carolina}, booktitle = {2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI)}, title = {In Silico Prediction of Cell Traction Forces}, year = {2020}, volume = {}, number = {}, pages = {877-881}, keywords = {Computer architecture;Neural networks;Uncertainty;Microprocessors;Force;Training;Entropy;Traction Force Microscopy;Deep Learning;Regression;Uncertainty;Bayesian Neural Network}, doi = {10.1109/ISBI45749.2020.9098359}, } - NeurIPS
 CoMIR: Contrastive Multimodal Image Representation for RegistrationNicolas Pielawski, Elisabeth Wetzer, Johan Öfverstedt, and 4 more authorsIn Advances in Neural Information Processing Systems, Mar 2020
CoMIR: Contrastive Multimodal Image Representation for RegistrationNicolas Pielawski, Elisabeth Wetzer, Johan Öfverstedt, and 4 more authorsIn Advances in Neural Information Processing Systems, Mar 2020We propose contrastive coding to learn shared, dense image representations, referred to as CoMIRs (Contrastive Multimodal Image Representations). CoMIRs enable the registration of multimodal images where existing registration methods often fail due to a lack of sufficiently similar image structures. CoMIRs reduce the multimodal registration problem to a monomodal one, in which general intensity-based, as well as feature-based, registration algorithms can be applied. The method involves training one neural network per modality on aligned images, using a contrastive loss based on noise-contrastive estimation (InfoNCE). Unlike other contrastive coding methods, used for, e.g., classification, our approach generates image-like representations that contain the information shared between modalities. We introduce a novel, hyperparameter-free modification to InfoNCE, to enforce rotational equivariance of the learnt representations, a property essential to the registration task. We assess the extent of achieved rotational equivariance and the stability of the representations with respect to weight initialization, training set, and hyperparameter settings, on a remote sensing dataset of RGB and near-infrared images. We evaluate the learnt representations through registration of a biomedical dataset of bright-field and second-harmonic generation microscopy images; two modalities with very little apparent correlation. The proposed approach based on CoMIRs significantly outperforms registration of representations created by GAN-based image-to-image translation, as well as a state-of-the-art, application-specific method which takes additional knowledge about the data into account. Code is available at: https://github.com/MIDA-group/CoMIR.
@inproceedings{comir2020, author = {Pielawski, Nicolas and Wetzer, Elisabeth and \"{O}fverstedt, Johan and Lu, Jiahao and W\"{a}hlby, Carolina and Lindblad, Joakim and Sladoje, Natasa}, booktitle = {Advances in Neural Information Processing Systems}, editor = {Larochelle, H. and Ranzato, M. and Hadsell, R. and Balcan, M.F. and Lin, H.}, pages = {18433--18444}, publisher = {Curran Associates, Inc.}, title = {CoMIR: Contrastive Multimodal Image Representation for Registration}, url = {https://proceedings.neurips.cc/paper_files/paper/2020/file/d6428eecbe0f7dff83fc607c5044b2b9-Paper.pdf}, volume = {33}, year = {2020}, }
2019
- Cytometry Part A
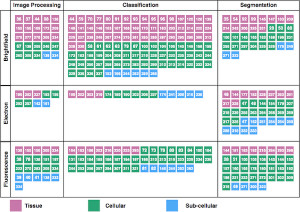 Deep Learning in Image Cytometry: A ReviewAnindya Gupta, Philip J. Harrison, Håkan Wieslander, and 9 more authorsCytometry Part A, Mar 2019
Deep Learning in Image Cytometry: A ReviewAnindya Gupta, Philip J. Harrison, Håkan Wieslander, and 9 more authorsCytometry Part A, Mar 2019Abstract Artificial intelligence, deep convolutional neural networks, and deep learning are all niche terms that are increasingly appearing in scientific presentations as well as in the general media. In this review, we focus on deep learning and how it is applied to microscopy image data of cells and tissue samples. Starting with an analogy to neuroscience, we aim to give the reader an overview of the key concepts of neural networks, and an understanding of how deep learning differs from more classical approaches for extracting information from image data. We aim to increase the understanding of these methods, while highlighting considerations regarding input data requirements, computational resources, challenges, and limitations. We do not provide a full manual for applying these methods to your own data, but rather review previously published articles on deep learning in image cytometry, and guide the readers toward further reading on specific networks and methods, including new methods not yet applied to cytometry data. © 2018 The Authors. Cytometry Part A published by Wiley Periodicals, Inc. on behalf of International Society for Advancement of Cytometry.
@article{DLCytometry2019, author = {Gupta, Anindya and Harrison, Philip J. and Wieslander, Håkan and Pielawski, Nicolas and Kartasalo, Kimmo and Partel, Gabriele and Solorzano, Leslie and Suveer, Amit and Klemm, Anna H. and Spjuth, Ola and Sintorn, Ida-Maria and Wählby, Carolina}, title = {Deep Learning in Image Cytometry: A Review}, journal = {Cytometry Part A}, volume = {95}, number = {4}, pages = {366-380}, keywords = {biomedical image analysis, cell analysis, convolutional neural networks, deep learning, image cytometry, microscopy, machine learning}, doi = {https://doi.org/10.1002/cyto.a.23701}, url = {https://onlinelibrary.wiley.com/doi/abs/10.1002/cyto.a.23701}, eprint = {https://onlinelibrary.wiley.com/doi/pdf/10.1002/cyto.a.23701}, year = {2019}, }